circacompare
circacompare.Rdcircacompare performs a comparison between two rhythmic groups of data. It tests for rhythmicity and then fits a nonlinear model with parametrization to estimate and statistically support differences in mesor, amplitude, and phase between groups.
Usage
circacompare(
x,
col_time,
col_group,
col_outcome,
period = 24,
alpha_threshold = 0.05,
timeout_n = 10000,
control = list(),
weights = NULL,
suppress_all = FALSE
)Arguments
- x
data.frame. This is the data.frame which contains the rhythmic data for two groups in a tidy format.
- col_time
The name of the column within the data.frame, x, which contains time in hours at which the data were collected.
- col_group
The name of the column within the data.frame, x, which contains the grouping variable. This should only have two levels.
- col_outcome
The name of the column within the data.frame, x, which contains outcome measure of interest.
- period
The period of the rhythm. For circadian rhythms, leave this as the default value, 24.
- alpha_threshold
The level of alpha for which the presence of rhythmicity is considered. Default is 0.05.
- timeout_n
The upper limit for the model fitting attempts. Default is 10,000.
- control
list. Used to control the parameterization of the model.- weights
An optional numeric vector of (fixed) weights. When present, the objective function is weighted least squares.
- suppress_all
Logical. Set to
TRUEto avoid seeing errors or messages during model fitting procedure. Default isFALSE.
Examples
df <- make_data(phi1 = 6)
out <- circacompare(
x = df, col_time = "time", col_group = "group",
col_outcome = "measure"
)
out
#> $plot
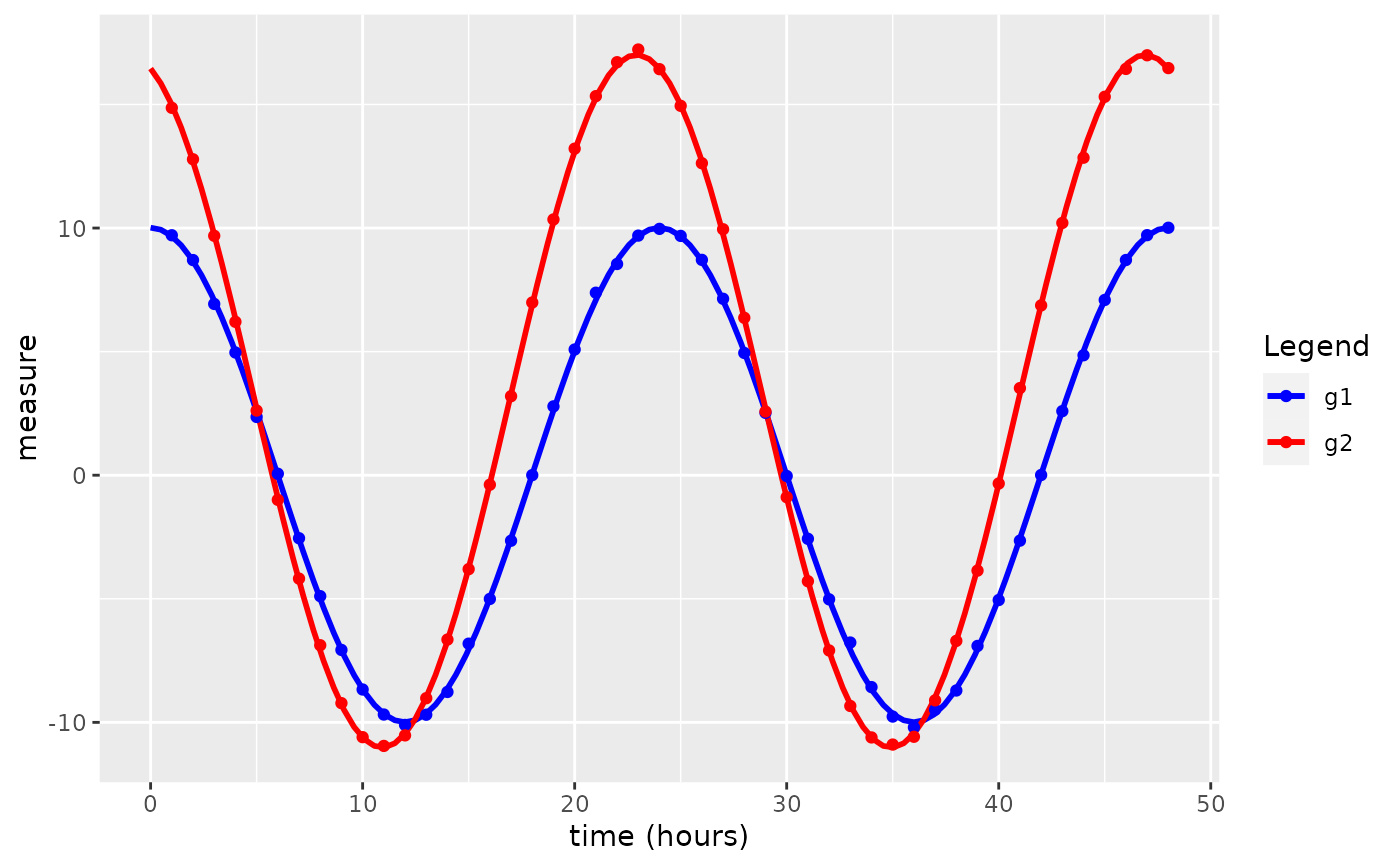 #>
#> $summary
#> parameter value
#> 1 Presence of rhythmicity (p-value) for g1 7.742499e-83
#> 2 Presence of rhythmicity (p-value) for g2 9.953293e-91
#> 3 g1 mesor estimate 1.089967e-02
#> 4 g2 mesor estimate 2.995890e+00
#> 5 Mesor difference estimate 2.984990e+00
#> 6 P-value for mesor difference 1.039187e-104
#> 7 g1 amplitude estimate 1.000437e+01
#> 8 g2 amplitude estimate 1.401390e+01
#> 9 Amplitude difference estimate 4.009528e+00
#> 10 P-value for amplitude difference 1.043910e-102
#> 11 g1 peak time hours 2.399305e+01
#> 12 g2 peak time hours 2.292035e+01
#> 13 Phase difference estimate -1.072698e+00
#> 14 P-value for difference in phase 2.308806e-94
#> 15 Shared period estimate 2.400000e+01
#>
#> $fit
#> Nonlinear regression model
#> model: measure ~ (k + k1 * x_group) + ((alpha + alpha1 * x_group)) * cos((1/period) * time_r - ((phi + phi1 * x_group)))
#> data: x
#> k k1 alpha alpha1 phi phi1
#> 0.0109 2.9850 10.0044 4.0095 12.5646 -0.2808
#> residual sum-of-squares: 1.11
#>
#> Number of iterations to convergence: 6
#> Achieved convergence tolerance: 4.046e-08
#>
# with sample weights (arbitrary weights for demonstration)
sw <- runif(n = nrow(df))
out2 <- circacompare(
x = df, col_time = "time", col_group = "group",
col_outcome = "measure", weights = sw
)
out2
#> $plot
#>
#> $summary
#> parameter value
#> 1 Presence of rhythmicity (p-value) for g1 7.742499e-83
#> 2 Presence of rhythmicity (p-value) for g2 9.953293e-91
#> 3 g1 mesor estimate 1.089967e-02
#> 4 g2 mesor estimate 2.995890e+00
#> 5 Mesor difference estimate 2.984990e+00
#> 6 P-value for mesor difference 1.039187e-104
#> 7 g1 amplitude estimate 1.000437e+01
#> 8 g2 amplitude estimate 1.401390e+01
#> 9 Amplitude difference estimate 4.009528e+00
#> 10 P-value for amplitude difference 1.043910e-102
#> 11 g1 peak time hours 2.399305e+01
#> 12 g2 peak time hours 2.292035e+01
#> 13 Phase difference estimate -1.072698e+00
#> 14 P-value for difference in phase 2.308806e-94
#> 15 Shared period estimate 2.400000e+01
#>
#> $fit
#> Nonlinear regression model
#> model: measure ~ (k + k1 * x_group) + ((alpha + alpha1 * x_group)) * cos((1/period) * time_r - ((phi + phi1 * x_group)))
#> data: x
#> k k1 alpha alpha1 phi phi1
#> 0.0109 2.9850 10.0044 4.0095 12.5646 -0.2808
#> residual sum-of-squares: 1.11
#>
#> Number of iterations to convergence: 6
#> Achieved convergence tolerance: 4.046e-08
#>
# with sample weights (arbitrary weights for demonstration)
sw <- runif(n = nrow(df))
out2 <- circacompare(
x = df, col_time = "time", col_group = "group",
col_outcome = "measure", weights = sw
)
out2
#> $plot
 #>
#> $summary
#> parameter value
#> 1 Presence of rhythmicity (p-value) for g1 3.989106e-85
#> 2 Presence of rhythmicity (p-value) for g2 7.489870e-92
#> 3 g1 mesor estimate -1.406791e-02
#> 4 g2 mesor estimate 2.989707e+00
#> 5 Mesor difference estimate 3.003775e+00
#> 6 P-value for mesor difference 2.973994e-108
#> 7 g1 amplitude estimate 1.000604e+01
#> 8 g2 amplitude estimate 1.401206e+01
#> 9 Amplitude difference estimate 4.006026e+00
#> 10 P-value for amplitude difference 7.126206e-106
#> 11 g1 peak time hours 2.399457e+01
#> 12 g2 peak time hours 2.291459e+01
#> 13 Phase difference estimate -1.079979e+00
#> 14 P-value for difference in phase 2.858438e-97
#> 15 Shared period estimate 2.400000e+01
#>
#> $fit
#> Nonlinear regression model
#> model: measure ~ (k + k1 * x_group) + ((alpha + alpha1 * x_group)) * cos((1/period) * time_r - ((phi + phi1 * x_group)))
#> data: x
#> k k1 alpha alpha1 phi phi1
#> -0.014068 3.003775 10.006039 4.006026 -0.001421 -0.282738
#> weighted residual sum-of-squares: 0.4322
#>
#> Number of iterations to convergence: 5
#> Achieved convergence tolerance: 3.213e-07
#>
#>
#> $summary
#> parameter value
#> 1 Presence of rhythmicity (p-value) for g1 3.989106e-85
#> 2 Presence of rhythmicity (p-value) for g2 7.489870e-92
#> 3 g1 mesor estimate -1.406791e-02
#> 4 g2 mesor estimate 2.989707e+00
#> 5 Mesor difference estimate 3.003775e+00
#> 6 P-value for mesor difference 2.973994e-108
#> 7 g1 amplitude estimate 1.000604e+01
#> 8 g2 amplitude estimate 1.401206e+01
#> 9 Amplitude difference estimate 4.006026e+00
#> 10 P-value for amplitude difference 7.126206e-106
#> 11 g1 peak time hours 2.399457e+01
#> 12 g2 peak time hours 2.291459e+01
#> 13 Phase difference estimate -1.079979e+00
#> 14 P-value for difference in phase 2.858438e-97
#> 15 Shared period estimate 2.400000e+01
#>
#> $fit
#> Nonlinear regression model
#> model: measure ~ (k + k1 * x_group) + ((alpha + alpha1 * x_group)) * cos((1/period) * time_r - ((phi + phi1 * x_group)))
#> data: x
#> k k1 alpha alpha1 phi phi1
#> -0.014068 3.003775 10.006039 4.006026 -0.001421 -0.282738
#> weighted residual sum-of-squares: 0.4322
#>
#> Number of iterations to convergence: 5
#> Achieved convergence tolerance: 3.213e-07
#>